Prediction of ligand binding sites using improved blind docking method with a Machine Learning-Based scoring function - ScienceDirect

Fully Blind Docking at the Atomic Level for Protein-Peptide Complex Structure Prediction. | Semantic Scholar

Fully Blind Docking at the Atomic Level for Protein-Peptide Complex Structure Prediction - ScienceDirect
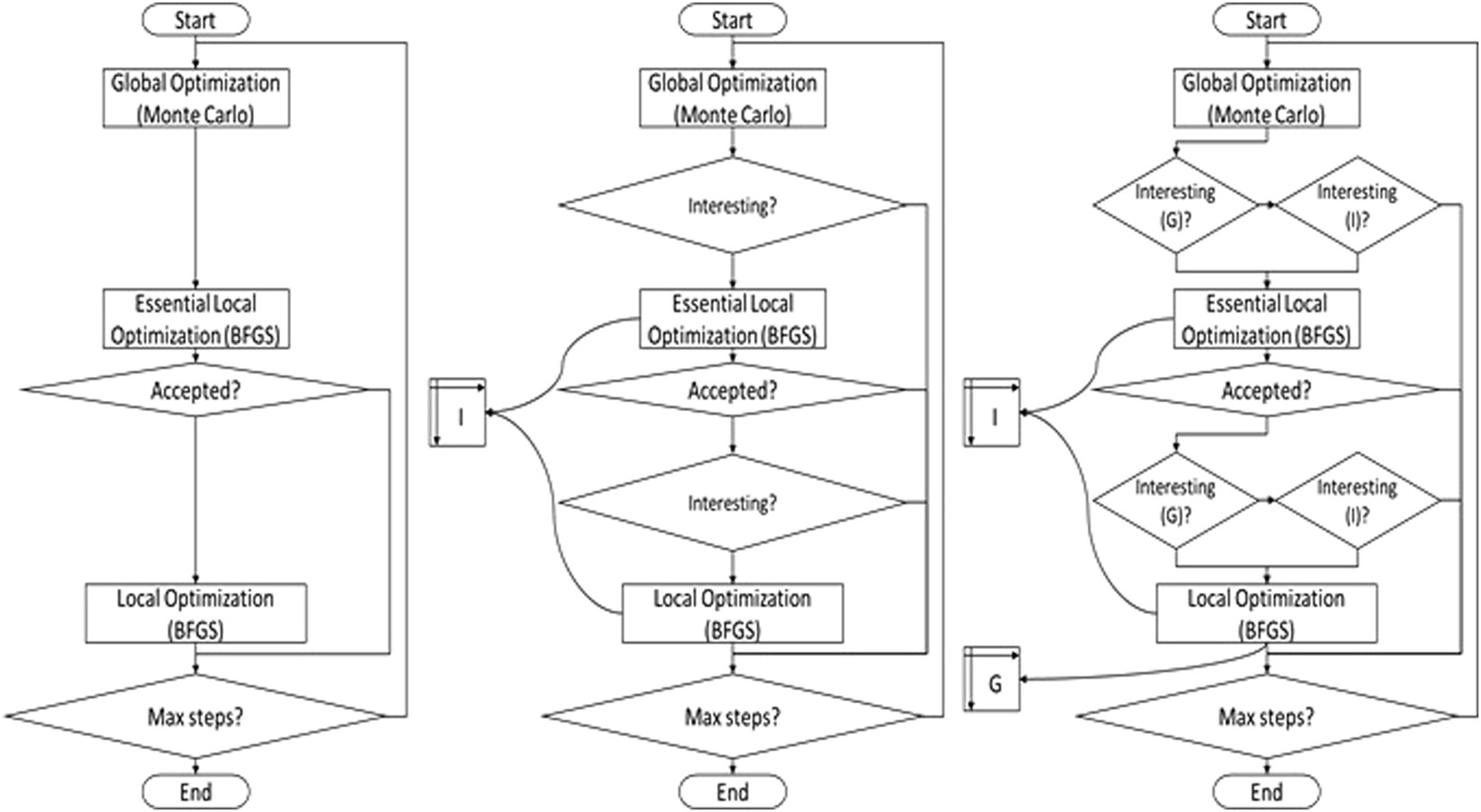
Protein-Ligand Blind Docking Using QuickVina-W With Inter-Process Spatio-Temporal Integration | Scientific Reports

Improving accuracy and efficiency of blind protein-ligand docking by focusing on predicted binding sites. - Abstract - Europe PMC
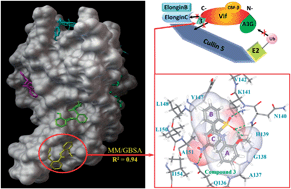
Exploring the binding mode of HIV-1 Vif inhibitors by blind docking, molecular dynamics and MM/GBSA - RSC Advances (RSC Publishing)

Site mapping and small molecule blind docking reveal a possible target site on the SARS-CoV-2 main protease dimer interface - ScienceDirect

Blind docking at exhaustiveness 32 (a) Different binding conformations... | Download Scientific Diagram
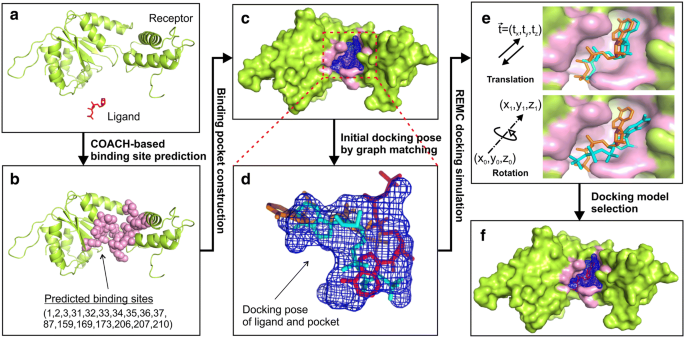
EDock: blind protein–ligand docking by replica-exchange monte carlo simulation | Journal of Cheminformatics | Full Text

Fully Blind Docking at the Atomic Level for Protein-Peptide Complex Structure Prediction. | Semantic Scholar
Improving accuracy and efficiency of blind protein-ligand docking by focusing on predicted binding sites


![PDF] DSDP: A Blind Docking Strategy Accelerated by GPUs | Semantic Scholar PDF] DSDP: A Blind Docking Strategy Accelerated by GPUs | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/b3f9ed5db9a0280d8c604ce18337bae8587c37d1/4-Figure1-1.png)